A Typical Trna Molecule Is Blank______ Bases Long.
Breaking News Today
Mar 13, 2025 · 8 min read
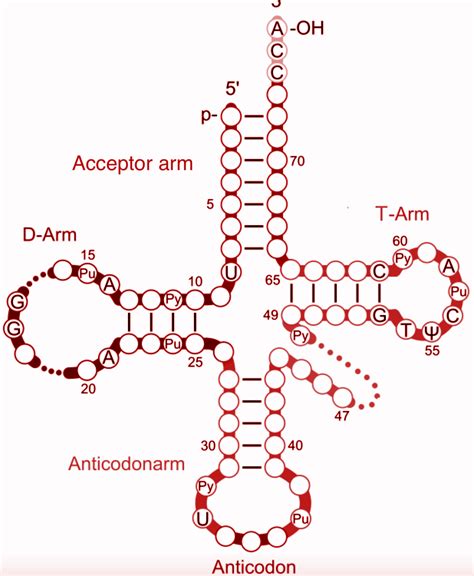
Table of Contents
A Typical tRNA Molecule is Approximately 75-90 Bases Long: A Deep Dive into Transfer RNA Structure and Function
A typical tRNA molecule is 75-90 bases long. This seemingly simple statement belies the incredible complexity and crucial role these small RNA molecules play in the fundamental process of protein synthesis. Understanding the structure and function of tRNA is essential to grasping the intricacies of molecular biology and the central dogma of life: DNA to RNA to protein. This article will explore the typical length of tRNA molecules, delve into the details of their secondary and tertiary structure, examine their key functional components, and highlight the significance of their size and shape in their overall function.
The Importance of tRNA Length and Structure
The relatively short length of tRNA, ranging from approximately 75 to 90 nucleotides, is not arbitrary. This specific size is precisely tailored to accommodate the molecule's complex three-dimensional structure and its vital roles in translation. The length allows for the formation of characteristic secondary and tertiary structural features crucial for tRNA function. These features are essential for:
-
Amino acid attachment: tRNA molecules act as adaptors, carrying specific amino acids to the ribosome based on the mRNA codon sequence. The 3' end of the tRNA molecule contains a CCA sequence, which is the site of amino acid attachment. The length of the tRNA ensures this critical region is properly positioned and accessible.
-
Codon recognition: The anticodon loop, a critical structural component, is responsible for recognizing and binding to the complementary codon on the mRNA molecule. The overall length and structure of the tRNA molecule contribute to the precise positioning and accessibility of this anticodon.
-
Ribosome interaction: tRNA molecules interact extensively with the ribosome, the protein synthesis machinery. The size and shape of the tRNA molecule are crucial for its proper binding and positioning within the ribosome's A, P, and E sites.
The Cloverleaf Secondary Structure: A Key Architectural Feature
tRNA molecules exhibit a characteristic cloverleaf secondary structure. This structure, while not perfectly representing the three-dimensional shape, provides a simplified and useful model for understanding the organization of the molecule. The cloverleaf structure is formed by base pairing within the tRNA sequence, creating several distinct arms or loops:
-
Acceptor stem: This is the stem at the 3' end of the tRNA molecule. The acceptor stem ends with the conserved CCA sequence, where the amino acid is attached. The length of this stem is relatively consistent across various tRNA species.
-
D arm: This arm contains dihydrouridine (D) residues, which are important for tRNA folding and interaction with the ribosome. The length and sequence of the D arm contribute to the overall stability and shape of the molecule.
-
TψC arm: This arm contains the TψC sequence (thymine-pseudouridine-cytidine). This sequence, along with the D arm, plays a critical role in recognition by aminoacyl-tRNA synthetases, enzymes that attach the correct amino acid to the tRNA. Variations in the length and sequence of this arm can influence tRNA specificity.
-
Anticodon arm: This crucial arm contains the anticodon, a three-nucleotide sequence that is complementary to a specific mRNA codon. The length of the anticodon arm and the exact position of the anticodon triplet are meticulously regulated, ensuring accurate codon recognition. The anticodon loop is often slightly longer than a simple hairpin loop, contributing to the accessibility and flexibility of the anticodon during codon-anticodon interactions.
-
Variable arm: This arm is the most variable region in tRNA molecules, both in terms of length and sequence. The variable arm's length significantly varies between different tRNA species, adding to the overall diversity of tRNA structures. This variability can contribute to the specificity of tRNA interactions with other molecules or within particular cellular compartments.
The L-Shaped Tertiary Structure: A Functional Masterpiece
While the cloverleaf model is useful for understanding the secondary structure, it doesn’t fully represent the three-dimensional conformation of tRNA molecules. In reality, tRNA adopts a characteristic L-shaped tertiary structure. This three-dimensional folding is achieved through tertiary interactions between various regions of the molecule, stabilized by hydrogen bonds and base stacking. The L-shape is vital for tRNA function, precisely orienting the acceptor stem and the anticodon loop for efficient interaction with the ribosome and mRNA.
The tertiary structure is stabilized by several interactions, including:
-
Base stacking: The planar aromatic bases stack upon one another, contributing to the stability and compactness of the L-shape.
-
Hydrogen bonds: Hydrogen bonds between specific bases further stabilize the tertiary structure, ensuring a precise three-dimensional arrangement.
-
Modified bases: Numerous modified bases are found in tRNA molecules, which enhance stability and influence the tertiary structure and function. The exact types and locations of these modified bases contribute to the diversity in tRNA conformations.
The L-shaped structure brings the acceptor stem (carrying the amino acid) and the anticodon loop (interacting with mRNA) into close proximity, facilitating the efficient transfer of amino acids during translation. This proximity is essential for the precise and efficient delivery of the correct amino acid to the growing polypeptide chain. The arrangement also enhances the interaction with various factors within the ribosome.
Aminoacyl-tRNA Synthetases: The Key Players in tRNA Charging
The accurate attachment of the correct amino acid to its cognate tRNA is a crucial step in protein synthesis. This process, called aminoacylation or charging, is catalyzed by aminoacyl-tRNA synthetases. These enzymes recognize specific tRNA molecules based on their secondary and tertiary structures, ensuring that each tRNA is loaded with the appropriate amino acid.
The recognition of tRNA by aminoacyl-tRNA synthetases involves multiple interactions between the enzyme and various regions of the tRNA molecule, including the acceptor stem, the D arm, and the TψC arm. The specific interactions are dependent on the particular tRNA molecule and its corresponding aminoacyl-tRNA synthetase. The overall length and structural features of the tRNA contribute to its unique identity and accurate recognition.
Variations in tRNA structure, such as the length and sequence of the variable arm and the presence of modified bases, can contribute to the specificity of tRNA recognition by aminoacyl-tRNA synthetases. This fine-tuning ensures that the appropriate amino acid is linked to the correct tRNA, preventing errors during protein synthesis.
Wobble Hypothesis and Codon-Anticodon Interactions
The anticodon loop of tRNA contains a triplet of nucleotides (the anticodon) that base pairs with the complementary codon on the mRNA molecule during translation. The interaction between the codon and anticodon is crucial for accurate protein synthesis. The wobble hypothesis explains how a single tRNA can recognize multiple codons encoding the same amino acid. This flexibility is accommodated by non-canonical base pairing at the third position (3' end) of the codon. The length and flexibility of the anticodon loop allow for this wobble base pairing, increasing the efficiency of translation by reducing the requirement for a unique tRNA for each codon.
The overall length of the tRNA molecule, particularly the length and structural features of the anticodon arm, influences the flexibility and accessibility of the anticodon during codon-anticodon interactions. Precise base pairing in the first two positions of the codon is usually maintained for accuracy, while some flexibility is permitted at the third position due to the inherent wobble.
The Role of Modified Nucleosides in tRNA Function
tRNA molecules often contain modified nucleosides, which are derived from the standard four nucleotides (adenine, guanine, cytosine, and uracil). These modifications contribute significantly to tRNA structure, stability, and function. These modifications can influence:
-
Tertiary structure: Modified nucleosides can influence the folding and stability of the L-shaped tertiary structure.
-
Codon-anticodon interactions: Modifications can affect the fidelity of codon-anticodon base pairing.
-
Recognition by aminoacyl-tRNA synthetases: Modified nucleosides can play a role in the recognition of tRNA by the appropriate synthetase.
-
Ribosome interactions: Modifications can influence the interactions between tRNA and the ribosome.
The number and types of modified nucleosides vary widely among different tRNA species, contributing to the diversity and specificity of tRNA function. The presence of these modifications often reflects the organism, cell type, and even specific physiological conditions.
Conclusion: The Exquisite Precision of tRNA Structure and Function
The statement that a typical tRNA molecule is approximately 75-90 bases long is a starting point for understanding the remarkable complexity and precision of these crucial molecules. The specific length, coupled with the intricate secondary and tertiary structures, ensures their ability to accurately perform their multifaceted roles in protein synthesis. The interaction between the tRNA molecule and the cellular machinery, the interplay between modified bases and their structural contributions, and the precise details of codon-anticodon pairing all highlight the exquisite precision and evolutionary optimization of tRNA structure and function. It is this detailed arrangement, honed by millions of years of evolution, that underpins the fundamental process of life itself. Further research continues to unveil the intricate details of tRNA function, highlighting its significance in cellular processes and potential implications for diverse fields of study.
Latest Posts
Latest Posts
-
Emotional Abuse Is Less Harmful Than Physical Abuse
May 09, 2025
-
Combustion Begins When A Fuel Is Heated To Its
May 09, 2025
-
Alcohol And Its Effects On The Body Worksheet Answers
May 09, 2025
-
The Four Principles That Guide Assistive Technology
May 09, 2025
-
The Team Will Play First Game On Saturday
May 09, 2025
Related Post
Thank you for visiting our website which covers about A Typical Trna Molecule Is Blank______ Bases Long. . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.