Select The Five Major Mechanisms Of Antimicrobial Resistance
Breaking News Today
Apr 01, 2025 · 5 min read
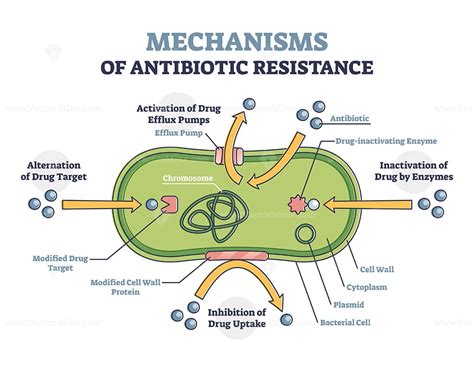
Table of Contents
The Five Major Mechanisms of Antimicrobial Resistance: A Deep Dive
Antimicrobial resistance (AMR) is a global health crisis, threatening our ability to treat common infections with antibiotics, antifungals, antivirals, and antiparasitic drugs. Understanding the mechanisms driving this resistance is crucial for developing effective strategies to combat it. This article delves into the five major mechanisms of antimicrobial resistance, explaining how they work and their implications for public health.
1. Target Modification: Altering the Antimicrobial's Binding Site
One of the most prevalent mechanisms of AMR involves altering the target site of the antimicrobial agent. This means the microorganism modifies the molecular structure of the drug's target, preventing the drug from binding and exerting its effect. Let's examine specific examples:
Target Modification in Bacteria:
-
Ribosomal alterations: Many antibiotics target bacterial ribosomes, essential for protein synthesis. Mutations in ribosomal proteins or ribosomal RNA can reduce the antibiotic's binding affinity, leading to resistance. This is a common mechanism for resistance to aminoglycosides, tetracyclines, and macrolides. For example, mutations in the 23S rRNA gene are frequently implicated in macrolide resistance.
-
Enzyme inactivation: Some antibiotics, like β-lactams (penicillins, cephalosporins), inhibit bacterial enzymes involved in cell wall synthesis. Bacteria can produce enzymes, such as β-lactamases, that inactivate these antibiotics by hydrolyzing the β-lactam ring. Extended-spectrum β-lactamases (ESBLs) and carbapenemases are particularly concerning as they can inactivate a broad range of β-lactams, including carbapenems, which are often used as last-resort antibiotics.
-
DNA gyrase mutations: Quinolones target DNA gyrase and topoisomerase IV, enzymes essential for DNA replication in bacteria. Mutations in these enzymes can reduce their affinity for quinolones, leading to resistance.
Target Modification in Other Microorganisms:
Target modification is not limited to bacteria. For example, resistance to antiviral drugs can occur through mutations in viral enzymes or receptors that the drug targets. Similarly, changes in the structure of fungal cell wall components can reduce the effectiveness of antifungal drugs.
2. Efflux Pumps: Pumping Antibiotics Out
Efflux pumps are transmembrane proteins that actively transport antimicrobial agents out of the bacterial cell. These pumps are naturally present in many bacteria, but their expression can be increased through mutations or regulatory changes, resulting in enhanced resistance.
How Efflux Pumps Work:
Efflux pumps function by binding to the antimicrobial, using energy (often from proton motive force) to move the drug from the cytoplasm across the cell membrane and out of the cell. A single efflux pump can often expel a broad range of structurally diverse antibiotics, contributing to multidrug resistance (MDR).
Clinical Significance of Efflux Pumps:
Overexpression of efflux pumps is a significant contributor to resistance in many clinically relevant bacteria, including Pseudomonas aeruginosa, Staphylococcus aureus, and Enterobacteriaceae. This mechanism often leads to resistance against multiple classes of antibiotics, making treatment challenging.
3. Enzyme Inactivation: Directly Neutralizing the Drug
As mentioned earlier in the context of β-lactamases, enzyme inactivation is a major mechanism of resistance. This involves the production of enzymes that chemically modify or degrade the antimicrobial agent, rendering it ineffective.
Examples of Enzyme Inactivation:
-
β-Lactamase Production (as discussed above): This remains a critical mechanism of resistance to β-lactam antibiotics. The variety of β-lactamases, their broad substrate profiles, and their capacity for transfer between bacteria pose serious challenges to treatment.
-
Aminoglycoside-Modifying Enzymes: These enzymes chemically modify aminoglycoside antibiotics, rendering them inactive. Modifications include acetylation, phosphorylation, and adenylation.
-
Chloramphenicol Acetyltransferase: This enzyme acetylates chloramphenicol, a protein synthesis inhibitor, inactivating it.
4. Reduced Permeability: Preventing Drug Entry
Some bacteria develop resistance by reducing the permeability of their outer membrane, preventing the antimicrobial agent from entering the cell in sufficient concentrations to exert its effect.
Outer Membrane Modifications:
Gram-negative bacteria possess an outer membrane that acts as a barrier to many antibiotics. Mutations affecting the porins (channels in the outer membrane) can decrease the permeability of the outer membrane, preventing antibiotics from reaching their intracellular targets. Alterations in lipopolysaccharide (LPS) structure can also contribute to reduced permeability.
Clinical Relevance of Reduced Permeability:
Reduced permeability is often observed in conjunction with other resistance mechanisms, creating a multi-layered defense against antibiotics. It is a particularly significant factor in the resistance of Gram-negative bacteria to many antibiotics.
5. Target Site Protection: Preventing Drug Access to the Target
This mechanism involves the production of proteins that physically protect the antimicrobial's target site, preventing the drug from binding.
Examples of Target Site Protection:
-
MecA in Staphylococcus aureus: Methicillin-resistant Staphylococcus aureus (MRSA) produces the penicillin-binding protein 2a (PBP2a), which has a low affinity for β-lactam antibiotics. This protein effectively protects the cell wall synthesis pathway from β-lactam inhibition.
-
Other Target Protection Mechanisms: Although less common than other mechanisms, variations in this strategy are observed in other pathogens. It involves proteins that bind to the antimicrobial or its target, hindering its activity.
Conclusion: The Complex Interplay of Resistance Mechanisms
The five major mechanisms of antimicrobial resistance – target modification, efflux pumps, enzyme inactivation, reduced permeability, and target site protection – often act in concert to create complex and highly resilient resistance profiles. Understanding these mechanisms is critical for developing novel antimicrobial strategies, including:
-
Developing new antibiotics that bypass existing resistance mechanisms: This includes designing drugs that target new bacterial pathways or that are less susceptible to existing resistance mechanisms.
-
Developing inhibitors of resistance mechanisms: This involves designing drugs that specifically target efflux pumps, β-lactamases, or other resistance-conferring enzymes.
-
Improving diagnostics to rapidly identify resistant organisms: This allows for timely treatment with appropriate antibiotics, minimizing the spread of resistance.
-
Implementing strategies to reduce the use and misuse of antibiotics: This is crucial to slowing the emergence and spread of resistance.
The fight against AMR requires a multifaceted approach involving researchers, clinicians, policymakers, and the public. By deepening our understanding of the intricate mechanisms driving resistance, we can work towards a future where antimicrobial agents remain effective in combating infectious diseases. The ongoing research into these mechanisms and the development of new strategies are vital to safeguarding global health.
Latest Posts
Latest Posts
-
Which Statement Accurately Describes Type 2 Diabetes
Apr 02, 2025
-
La Juventud Es La Etapa Cuando Nos Jubilamos
Apr 02, 2025
-
Practicing Good Manners In The Workplace Is Referred To As
Apr 02, 2025
-
Which Statement About Self Injury Is True
Apr 02, 2025
-
You Have Been Performing Multiple Provider Cpr And Using An Aed
Apr 02, 2025
Related Post
Thank you for visiting our website which covers about Select The Five Major Mechanisms Of Antimicrobial Resistance . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
