The Nucleotide Sequence In Mrna Is Determined By
Breaking News Today
Mar 19, 2025 · 6 min read
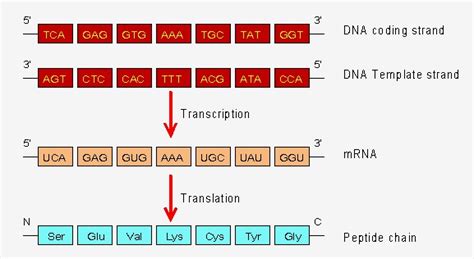
Table of Contents
The Nucleotide Sequence in mRNA is Determined By: A Deep Dive into Transcription and Beyond
The central dogma of molecular biology dictates the flow of genetic information: DNA → RNA → protein. Understanding how the nucleotide sequence in messenger RNA (mRNA) is determined is fundamental to grasping this process. This journey begins with DNA, the blueprint of life, and culminates in the synthesis of mRNA, a crucial intermediary carrying the genetic code to the ribosome for protein synthesis. This article will explore the intricate mechanisms underlying mRNA sequence determination, delving into the complexities of transcription, RNA processing, and the factors influencing the final mRNA product.
The Role of DNA: The Master Template
The nucleotide sequence in mRNA is fundamentally determined by the DNA sequence of the gene it transcribes. DNA, a double-stranded helix, carries the genetic information encoded in the sequence of its four nitrogenous bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Each gene within the DNA molecule represents a specific segment encoding a particular protein or functional RNA molecule. The process of transcription faithfully copies the nucleotide sequence of this gene into a complementary mRNA molecule.
The Transcription Process: A Step-by-Step Guide
Transcription, catalyzed by the enzyme RNA polymerase, is a multi-step process:
-
Initiation: RNA polymerase binds to a specific region of the DNA molecule called the promoter. Promoters are typically located upstream of the gene's coding sequence and contain specific DNA sequences recognized by RNA polymerase and other transcription factors. The binding of these factors initiates the unwinding of the DNA double helix, exposing the template strand.
-
Elongation: RNA polymerase moves along the template strand of DNA, synthesizing a complementary RNA molecule. The enzyme adds ribonucleotides (A, U, G, and C) to the growing RNA chain, following the base-pairing rules: A pairs with U (uracil replaces thymine in RNA), and G pairs with C. This process continues along the gene's coding sequence until the termination signal is reached.
-
Termination: Specific DNA sequences act as termination signals, signaling the end of transcription. These signals trigger the release of the newly synthesized mRNA molecule from the DNA template and RNA polymerase.
The Template Strand and the Coding Strand
It's crucial to understand the concept of the template strand and the coding strand. The template strand is the DNA strand that serves as the template for RNA synthesis. The RNA molecule synthesized is complementary to the template strand. The coding strand, also known as the non-template strand, has the same sequence as the mRNA molecule (except for uracil replacing thymine). Understanding this distinction is important for interpreting DNA sequences and predicting the resulting mRNA sequence.
Beyond Transcription: RNA Processing
The newly synthesized mRNA molecule is often not immediately ready for translation. In eukaryotic cells, it undergoes several processing steps before it can leave the nucleus and enter the cytoplasm for protein synthesis. These processing steps significantly influence the final mRNA nucleotide sequence.
Capping: Protecting the 5' End
The 5' end of the pre-mRNA molecule undergoes capping. A modified guanine nucleotide (7-methylguanosine) is added to the 5' end through a 5'-5' triphosphate linkage. This cap protects the mRNA from degradation and is crucial for ribosome binding during translation. While the cap itself doesn't alter the coding sequence, it is essential for the mRNA's stability and function.
Splicing: Removing Introns
Eukaryotic genes are characterized by the presence of introns and exons. Introns are non-coding sequences interspersed within the coding sequences (exons). Splicing is a process where introns are removed from the pre-mRNA molecule, and exons are joined together. This process is catalyzed by a complex called the spliceosome, which recognizes specific sequences at the intron-exon boundaries. Splicing significantly alters the nucleotide sequence of the pre-mRNA, removing the intron sequences and joining the exons to create a continuous coding sequence.
Polyadenylation: Adding a Poly(A) Tail
The 3' end of the pre-mRNA molecule is processed by adding a poly(A) tail. This tail consists of a string of adenine nucleotides (typically 100-200) and is added by the enzyme poly(A) polymerase. The poly(A) tail protects the mRNA from degradation, enhances its stability, and plays a role in its transport from the nucleus to the cytoplasm. Again, while not directly changing the coding sequence, it profoundly impacts mRNA lifespan and functionality.
Factors Influencing mRNA Sequence: Beyond the Basics
Several other factors can influence the final mRNA nucleotide sequence, including:
-
Alternative splicing: A single gene can generate multiple mRNA isoforms through alternative splicing. This involves selecting different combinations of exons during splicing, leading to the production of proteins with varying structures and functions. This dramatically increases the proteome complexity from a limited genome size.
-
RNA editing: Certain enzymes can modify the nucleotide sequence of the pre-mRNA molecule after transcription. This can involve the insertion, deletion, or modification of individual nucleotides, leading to changes in the protein's amino acid sequence.
-
RNA interference (RNAi): Small RNA molecules, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs), can regulate gene expression by binding to target mRNA molecules and either promoting their degradation or inhibiting their translation. Though not directly altering the sequence, this fundamentally alters the amount and therefore effective sequence available for translation.
-
Transcriptional regulation: The rate of transcription itself is controlled by various factors, including transcription factors, enhancers, and silencers. These regulatory elements influence the amount of mRNA produced from a given gene, ultimately impacting the overall representation of that gene's sequence in the cell. This impacts gene expression, meaning the final mRNA product may not be expressed consistently throughout all cells and stages of development.
Conclusion: A Dynamic Process
The determination of the nucleotide sequence in mRNA is not a static process but a dynamic and intricately regulated one. It starts with the DNA sequence, is faithfully transcribed into a pre-mRNA molecule, and is then subjected to various processing steps that significantly influence the final mRNA product. Alternative splicing, RNA editing, RNAi, and transcriptional regulation add layers of complexity, ensuring that the cell can fine-tune gene expression and adapt to changing conditions. Understanding these processes is crucial for comprehending the complexities of gene expression, protein synthesis, and the overall functioning of living organisms. The precise sequence, therefore, is a result of a complex interplay of molecular machinery and regulatory mechanisms ensuring the accurate and efficient expression of the genetic code. Further research continues to uncover the nuanced details of this fundamental biological process.
Latest Posts
Latest Posts
-
Appeasement Was A Popular Policy Because European Leaders
Mar 19, 2025
-
Wrasse Fish Black Sea Bass Info On Relationship
Mar 19, 2025
-
Chriss Views On Money And Governmental Authority
Mar 19, 2025
-
Removing An Organism From An Ecosystem
Mar 19, 2025
-
First Ten Elements On The Period Table
Mar 19, 2025
Related Post
Thank you for visiting our website which covers about The Nucleotide Sequence In Mrna Is Determined By . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
